Andy M. Lau
Structural Bioinformatician
University College London
Biography
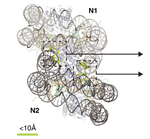
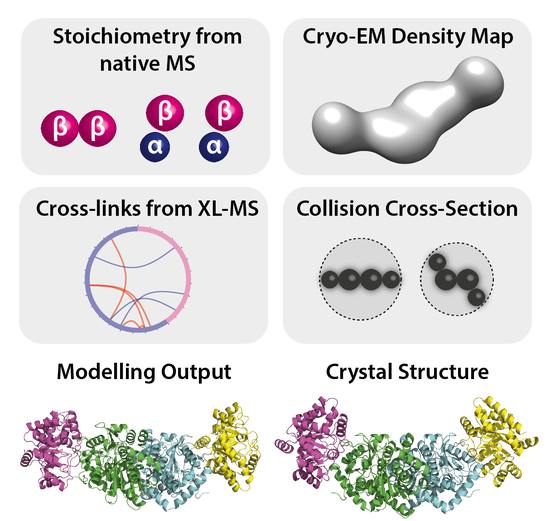
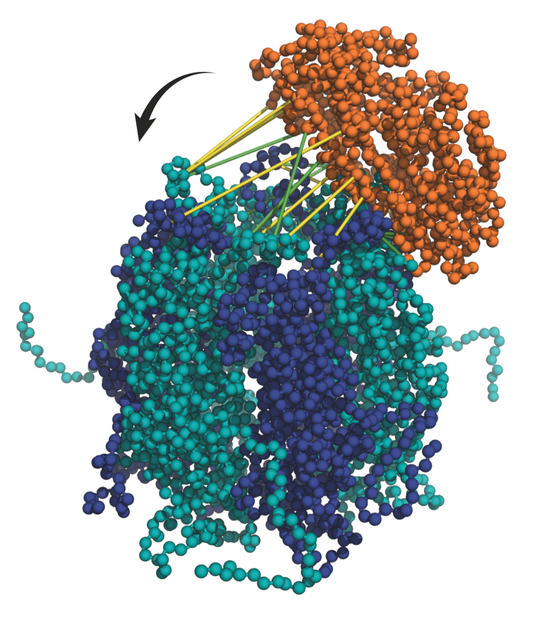
I am a Research Fellow currently based at University College London in the group of Prof. David Jones, with interests in predicting protein structure and behaviour using machine learning methods. My PhD at King’s College London in the group of Argyris Politis, developed methods of integrating data from structural mass spectrometry techniques (such as IM-MS, HDX-MS and XL-MS) together with data from X-ray crystallography and cryo-EM, with computational modelling to elucidate the conformational dynamics of heterogeneous biomolecules such as antibodies and multi-subunit complexes.
I developed Deuteros, software that performs analysis and visualisation of data from HDX-MS. My interests are in structural biology, particularly in protein structure, dynamics and understanding the interplay between their intricate conformations and functions.
Interests
- Machine learning
- Protein structure prediction
- Protein dynamics
Education
-
PhD in Structural Bioinformatics, 2019
King's College London
-
BSc in Biochemistry, 2015
King's College London